Ongoing Research Projects
This page briefly highlights some of the projects being worked on in the KLab. Further details about Randy Klabacka's focal projects can be found at Randy's personal website .
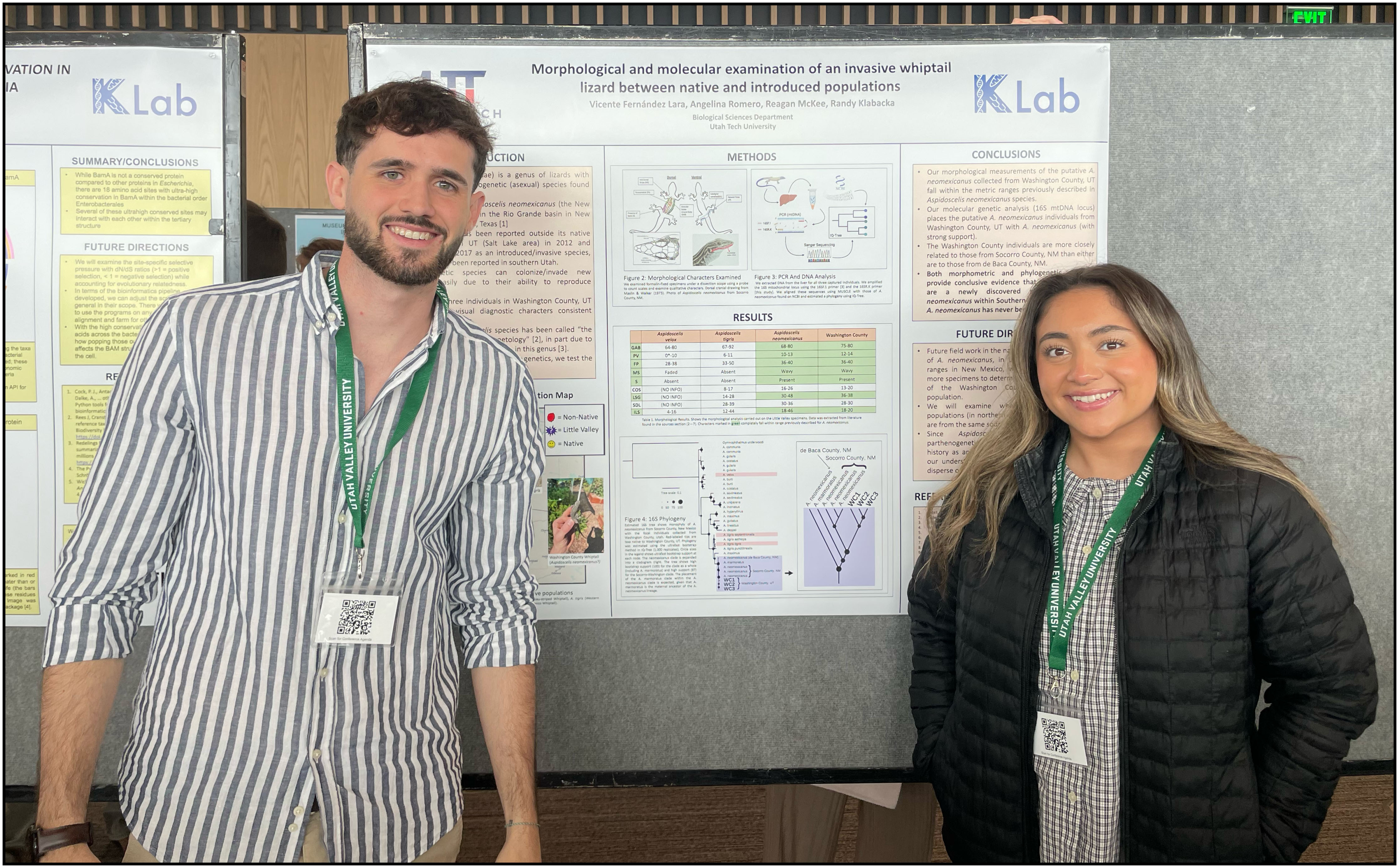
Conservation Phylogenetics: Discovery of a potential Non-native Lizard
The parthenogenic New Mexico whiptail lizard Aspidoscelis neomexicanus has established introduced populations in Northeastern Arizona and Northern Utah, but no populations were described outside of Salt Lake and Utah counties. Vicente Fernández Lara, Angelina Romero, and Reagan McKee discovered a new population in Southern Utah (Washington County). After preserving specimens and extracting DNA, Vicente, Angelina, and Reagan amplified and sequenced a mitochondrial gene to verify their species identity using phylogenetic inference.

Relationship of NGS read mapping success and phylo distance
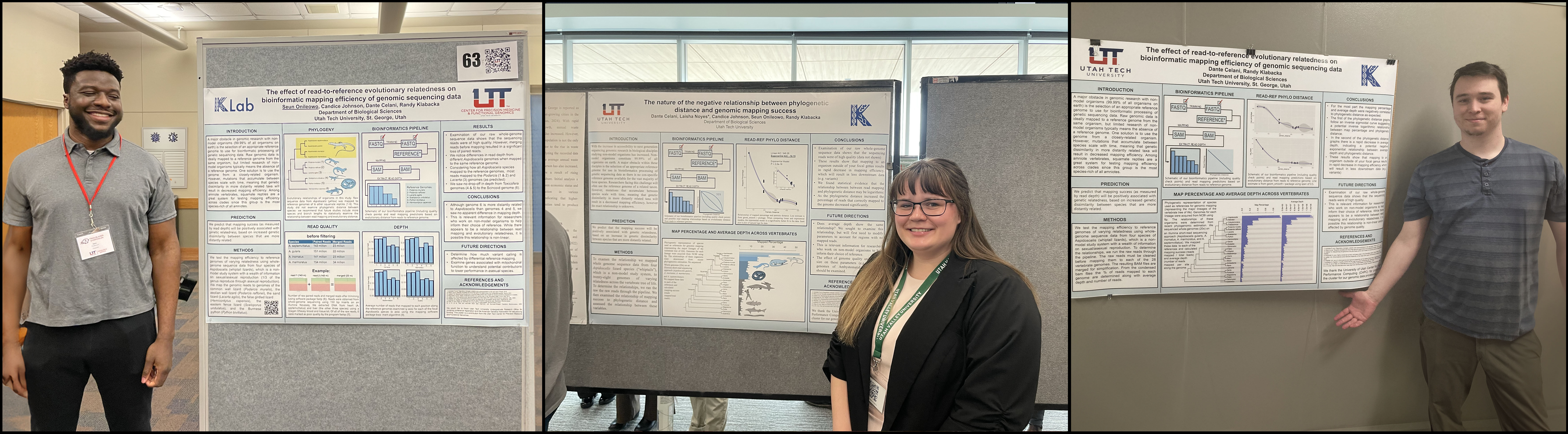
For non-model organisms, projects that use next-generation sequencing (NGS) data are challenging due to the lack of reference genomes available. This requires the mapping of sequence data to a species with a genome available or the sequencing and assembly of a reference-quality genome (the latter of which is sometimes financially and logistically unfeasible). Mapping is the step in bioinformatic data processing where NGS reads are aligned to a reference genome. It is understood that with increased evolutionary distance between the read and reference taxa the mapping success decreases, however the exact relationship of this decrease is not characterized. For this project, we map sequence data from four lizard species in the genus Aspidoscelis to reference genomes across the vertebrate tree of life and examine the relationship of mapping success to phylogenetic distance. Dante Celani led this project with help from Candice Johnson, Seun Onileowo, and Laisha Noyes are leading this project.
Freeze-resistant Protein Evolution
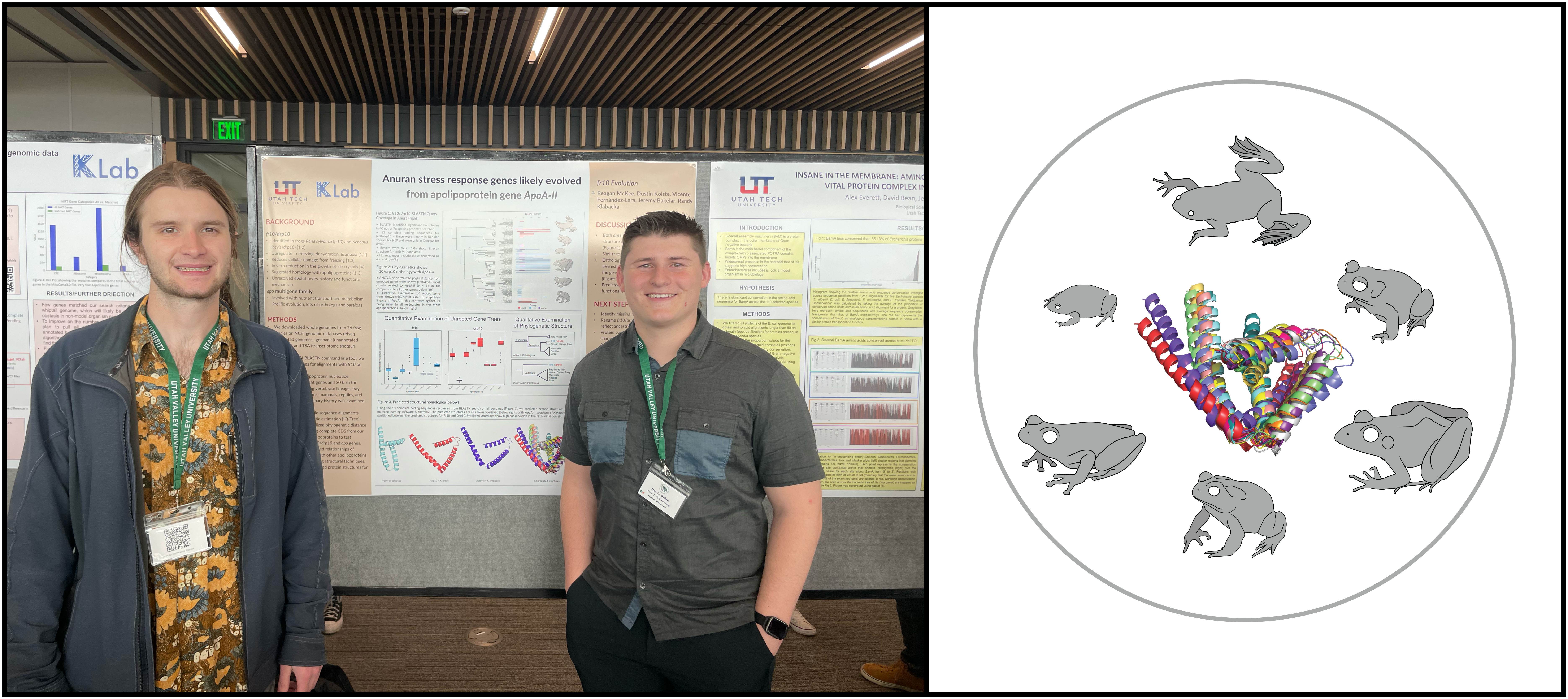
A small peptide found in a species of frog confers the ability to withstand freezing temperatures without ice crystals forming within the tissues. We are using a bioinformatic approach to seaerch for this gene in the genomes of other lineages across the frog tree of life. Preliminary examination of over 40 frog genomes/transcriptomes shows that fr10 is present in other Ranids, and it appears to be an apolipoprotein. Reagan McKee is leading this project with Dustin Kolste- they presented their work at UCUR 2024.
BamA and Potra Domain Diversity
BamA is a vital protein within the beta-barrel assembly machinery in gram-negative bacteria, and the domains (BamA and its associated potra domains) of this amino acid chain are potential targets of anti-microbial drugs. Understanding areas of high conservation that would be most likely to avoid nucleotide substitution provides candidate loci for drug development. We are examining the nucleotide diversity of this gene and levels of locus conservation across several levels of phylogenetic diversity. Preliminary examination reveals that several regions are completely conserved across the bacterial tree of life. Alex Everrett and David Bean are leading this project in collaboaration with Dr. Jeremy Bakelar. Check out their "Bam Machinery Conservation"

Computational Tool for Population-aware Ecotype Divergence
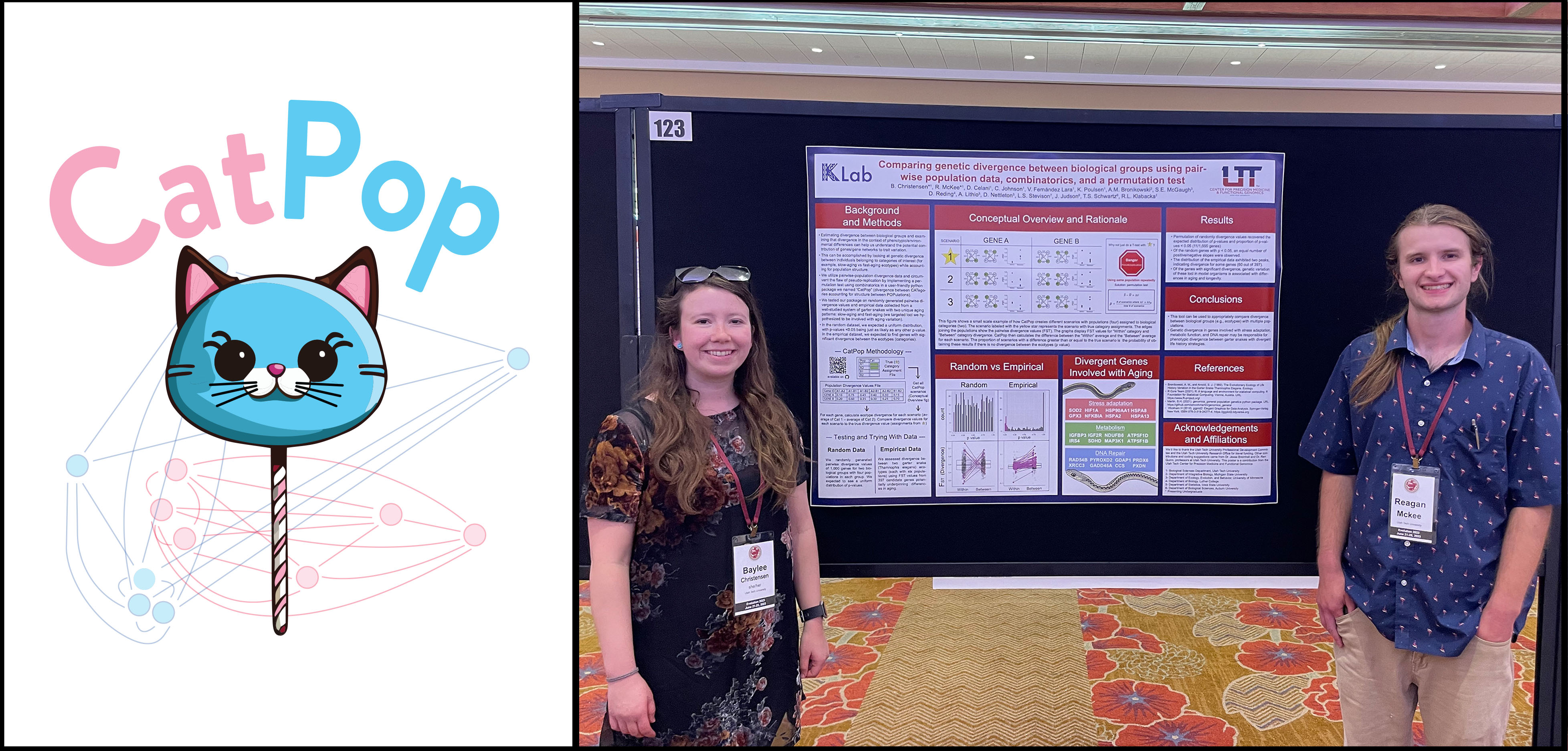
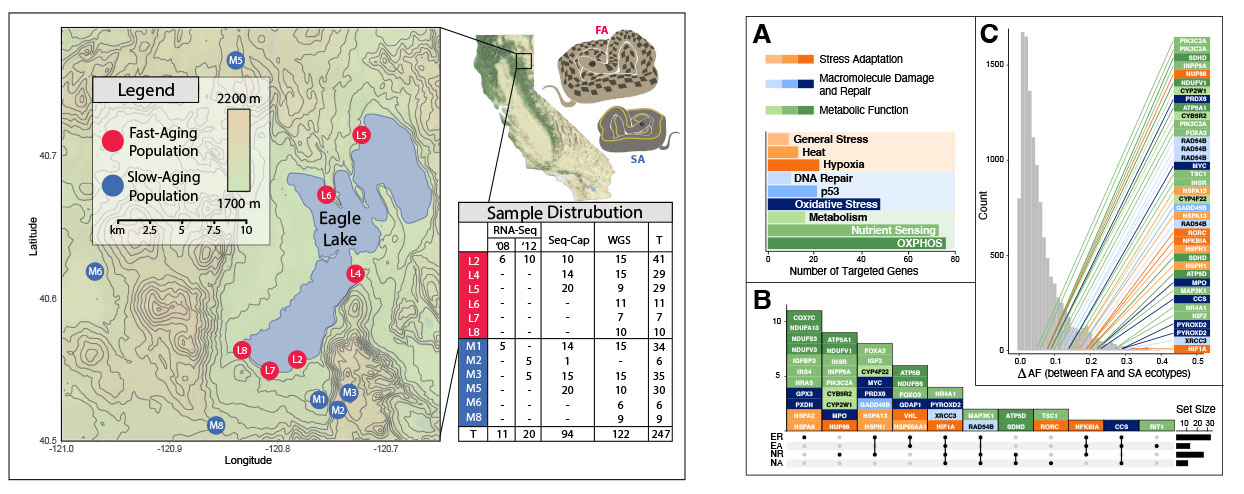
In biological study systems where multiple populations can be assigned to distinguishable categories (e.g., A and B), approaches to quantify genetic divergence between the categories should incorporate divergence between the populations themselves. To do this, calculations of divergence for pairwise population comparisons can be binned into “within” (comparisons of populations in the same category; e.g., A1–A2 and B1–B2) or “between” (comparisons of populations in different categories; e.g., A1–B1, A2–B2, A1–B2, A2–B1). However, this presents a potential pitfall of statistical pseudoreplication due to multiple measurements of the same population. To circumvent the flaw of pseudoreplication, we developed a software package (called "CatPop") that uses combinatorics to create all possible scenarios of population assignments to categories and perform a permutation test to identify genes where there is greater divergence in “between” comparisons compared to “within” comparisons. We validate CatPop’s statistical approach using a randomly generated dataset of divergence values in 8 populations with 1,000 genes. We then use CatPop to examine genetic divergence in an empirical dataset of 397 genes of interest from six garter snake populations assigned to two categories (a “fast-aging” ecotype and a “slow-aging” ecotype) and identify significant genes in several age-associated gene networks, including pathways of nutrient-sensing, DNA macromolecule damage/repair, and stress adaptation. Baylee Christensen, Reagan McKee, Dante Celani, and Candice Johnson led this project in collaboration with many other researchers.
Functional and Population Genomics of Divergent Garter Snake Ecotypes
Understanding the factors contributing to variation in life history traits like lifespan, growth rate, fecundity, and senescence across different species is crucial for comprehending the selection targets leading to divergence in these traits. While aging is a widespread phenomenon in the life history of many organisms, most studies focus on model organisms in laboratory settings. To identify genetic mechanisms influencing aging, it is essential to explore natural populations with aging variation. In the case of terrestrial garter snakes around Eagle Lake in Northeastern California, two ecotypes exhibit distinct life history strategies, with one aging faster than the other. Investigating gene networks associated with aging, I identified patterns in sequence variation in candidate genes between these ecotypes, including nonsynonymous variants with predicted functional impacts on gene products. This study builds on decades of research on these ecotypes, highlighting the genomic underpinnings of their ecological and physiological differences. This project is in its final stages of manuscript preparation for the journal Aging Cell
Mitonuclear Gene Variation in Whiptail Lizards
Whiptail lizards (genus Aspidoscelis) are unique in that roughly one-third of the genus is parthenogenic (reproduces primarily via asexual reproduction). These parthenogenetic species are the result of hybridization events between divergent sexual species, and the parthenogens exhibit lower endurance capacity compared to their sexual progenitors. To examine if the lower endurance could be due to mismatches in mitonuclear genes, we are examining the genetic variation in mitonuclear genes of four sexual Aspidoscelis. Dante Celani is leading this project.
Differential Gene Expression in Whiptails
Parthenogenetic lizards present a great model to understand the effects of selection and phenotypic plasticity in a non-recombining species with very little genetic variation. We sequenced transcriptomes from three populations of a parthenogenetic whiptail species (Aspidoscelis tesselatus) and will examine how gene expression in skeletal muscle tissue varies across the populations (which are located at different latidutidnal positions along the Rio Grande river basin in New Mexico, USA. Additionally, for one of the populations we collected samples from three different tissue types (skeletal muscle, liver, and heart) to examine differences in expression patterns across the tissues.
Mitonuclear Mismatching in Hybrid Parthenogens
Some instances of natural hybridization in reptiles led to the formation of new species that reproduce asexually (Barley et al., 2022). In these species, offspring are produced without male contribution (i.e., these species are all female). For these asexual lizards, their genomes are locked in a hybrid state (Vrijenhoek & Pfeiler, 2008; Warren et al., 2018) (i.e., no sexual recombination occurs to alter their genetic composition). Asexual whiptail lizards exhibit reduced mitochondrial respiration (Cullum, 1997;Klabacka et al., 2022). Explicit examination of the molecular mechanisms involved in mito-nuclear mismatch is needed to understand which genes are responsible for reduced organism health in hybrid species.
For this project we will bioinformatically examine the vast network of genes involved in mitochondrial respiration of hybrid asexual organisms (Aspidoscelis tesselatus) at three levels: (1) nucleotide sequence [DNA], (2) transcript sequence/abundance [RNA], and (3) amino acid sequence [protein]. Examination of these genes will help us understand (A) the specific genetic underpinnings of reduced mitochondrial respiration in hybrid asexual organisms, (B) whether hybrid asexual organisms can cope with mito-nuclear mismatch by “turning off” (silencing) or “turning down” (suppressing) genes that aren’t co-evolved with their mitochondria, and (C) the role of mito-nuclear interactions in speciation.
Prospective Research Projects
Mitonuclear Phylogenetics:
In Eukaryotic organisms, DNA resides inside both the nucleus and the mitochondria. Both of these genomes contain information that encodes cellular gene products (e.g., proteins and functional RNA). All of the gene products from the mitochondrial genome perform functions within the mitochondria, whereas the vast majority of gene products from the nuclear genome do not. However, a subset of gene products from the nuclear genome target the mitochondria; these gene products (from over 1,000 nuclear genes known as 'N-mt' genes) must interact with those of the mitochondrial genome. Incompatibilities between the gene products can result in decreased mitochondrial function, which can negatively affect cellular, organismal, and organismal performance.
In species with mitonuclear phylogenetic discordance (where the nuclear phylogeny doesn't match the mitochondrial phylogeny), it is common for the mitochondrial phylogeny to show more resolved, divergent lineages in contrast to the nuclear phylogeny's unresolved, admixed lineages. We are testing whether the evolutionary history of N-mt genes in such scenarios reflects that of the mitochondrial tree or the nuclear tree.
Life History Evolution
Females of the side-blotched lizard (Uta stansburiana) are hypothesized to have divergent life history strategies associated with color polymorphism. Orange-throated individuals lay large clutches of small eggs, whereas yellow- and blue-throated individuals lay small clutches of larger eggs. Differences in reproductive strategy are frequently associated with differences in aging (i.e., rate of senescence, longevity) and growth (e.g., growth rate, adult body size). We will test whether female throat color variation is associated with differences in growth rate and aging, and examine the genetic contribution to these life history traits.
Skeletomuscular Histology
Aspidoscelis lizards use their forelimbs and hindlimbs for completely different purposes (forelimbs for foraging and burrowing, hindlimbs for quick escapes). Because of this, their muscular physiology appears to be different. We will test the mitochondrial content of the different skeletal muscle tissue and examine the mitochondrial morphology using transmission electron microscopy.